4.3 Importing Data from Delimited Files
Knowing how to export data into delimited files, let us now see how to import data from delimited files.
4.3.1 Import .csv Files using read_csv()
To import .csv files, we can use the function read_csv() in the readr package, which is a sub-package of tidyverse. If you have already installed tidyverse, you can directly load the readr package.
After loading the readr package, you can try to import the data from “gm_small.csv” which we created in Section 4.2.3. Please make sure the .csv file is located in the folder “data” relative to the current working directory. Otherwise, you need to change the working directory accordingly using the methods introduced in Section 4.2.1.
library(readr)
gm_small <- read_csv("data/gm_small.csv")
#> Rows: 5 Columns: 4
#> ── Column specification ─────────────────────────────────────────
#> Delimiter: ","
#> chr (3): country, gender, continent
#> dbl (1): year
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.We can see that there is a message showing the Column specification during the import process. In particular, we see country is of type character, and both year and HDI are of type double. We can also check the value of gm_small and its structure.
gm_small
#> # A tibble: 5 × 4
#> country year gender continent
#> <chr> <dbl> <chr> <chr>
#> 1 Albania 2004 female Europe
#> 2 Andorra 2004 female <NA>
#> 3 United Arab Emirates 2004 female Asia
#> 4 Argentina 2004 female Americas
#> 5 Armenia 2004 female Asia
str(gm_small)
#> spc_tbl_ [5 × 4] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
#> $ country : chr [1:5] "Albania" "Andorra" "United Arab Emirates" "Argentina" ...
#> $ year : num [1:5] 2004 2004 2004 2004 2004
#> $ gender : chr [1:5] "female" "female" "female" "female" ...
#> $ continent: chr [1:5] "Europe" NA "Asia" "Americas" ...
#> - attr(*, "spec")=
#> .. cols(
#> .. country = col_character(),
#> .. year = col_double(),
#> .. gender = col_character(),
#> .. continent = col_character()
#> .. )
#> - attr(*, "problems")=<externalptr>We can see that the tibble gm_small is generated along with the correct column types. In order to introduce the various options associated with read_csv() function, let’s move on to the topic of inline .csv files next.
4.3.2 Read Inline .csv Files
The read_csv() function not only can read files into R, it also accept inline input as its argument. While the inline input may not be commonly used in practice, it is particularly useful for learning how to use the function, and be able to handle various importing cases when dealing with real life datasets. Let’s see an example.
read_csv("x,y,z
1,3,5
2,4,6")
#> Rows: 2 Columns: 3
#> ── Column specification ─────────────────────────────────────────
#> Delimiter: ","
#> dbl (3): x, y, z
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
#> # A tibble: 2 × 3
#> x y z
#> <dbl> <dbl> <dbl>
#> 1 1 3 5
#> 2 2 4 6This inline input example is equivalent to reading a .csv file with contents identical to those in quotes. You can see that a tibble with size of 2 by 3 is generated with the column names being x, y and z. From the argument, we can see that by default, the first row of the input data will be interpreted as the column names.
a. No column names
If the first row of the input data doesn’t correspond to the variable names, you need to set col_names = FALSE as an additional argument in read_csv().
read_csv("x,y,z
1,3,5
2,4,6", col_names = FALSE)
#> Rows: 3 Columns: 3
#> ── Column specification ─────────────────────────────────────────
#> Delimiter: ","
#> chr (3): X1, X2, X3
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
#> # A tibble: 3 × 3
#> X1 X2 X3
#> <chr> <chr> <chr>
#> 1 x y z
#> 2 1 3 5
#> 3 2 4 6Now, a tibble of 3 rows and 3 columns was generated, with the column names being X1, X2, and X3. Note that these are the default naming conventions in the function when you don’t supply the column names in the file. Another thing worth mentioning is that all three variables are of character types, due to the fact that there are character values for all variables (“x”, “y”, and “z”).
b. Skip the first few lines
Sometimes, the first few lines of your data file may be descriptions of the data, which you want to skip when importing into R. We can set the skip argument in the read_csv() function to skip a certain number of lines.
read_csv("The first line
The second line
The third line
x,y,z
1,3,5",
skip = 3)
#> Rows: 1 Columns: 3
#> ── Column specification ─────────────────────────────────────────
#> Delimiter: ","
#> dbl (3): x, y, z
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
#> # A tibble: 1 × 3
#> x y z
#> <dbl> <dbl> <dbl>
#> 1 1 3 5It is clear from the result that the first 3 lines of the input data are skipped.
c. Skip the comments
Another useful argument in cases when we have comments in the data file is the comment argument, which tells R to skip all text after the string specified in the comment argument.
read_csv("x,y,z #variable names
1,3,5 #the first observation
2,4,6 #the second observation",
comment = "#")
#> Rows: 2 Columns: 3
#> ── Column specification ─────────────────────────────────────────
#> Delimiter: ","
#> dbl (3): x, y, z
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
#> # A tibble: 2 × 3
#> x y z
#> <dbl> <dbl> <dbl>
#> 1 1 3 5
#> 2 2 4 64.3.3 Handling Missing Values
In many real data sets, we may have missing values. You may recall that R uses NA to represent the missing values. If the data set was prepared by an R user, it probably already uses NA to represent all missing values. In this case, life is easy since read_csv() will automatically interpret all NAs as missing values.
read_csv("x,y,z
999,3,5
NA,-999,6")
#> Rows: 2 Columns: 3
#> ── Column specification ─────────────────────────────────────────
#> Delimiter: ","
#> dbl (3): x, y, z
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
#> # A tibble: 2 × 3
#> x y z
#> <dbl> <dbl> <dbl>
#> 1 999 3 5
#> 2 NA -999 6In a typical application, however, the person who prepared the data may use other strings to represent missing values. For example, if 999 and -999 are used as the indicators for missing values, you can set the argument na to be the vector containing those values. Note that NA will still be interpreted as missing values automatically.
read_csv("x,y,z
999,3, yes
NA,-999, no
12, 12, NA",
na = c("999", "-999"))
#> Rows: 3 Columns: 3
#> ── Column specification ─────────────────────────────────────────
#> Delimiter: ","
#> chr (2): x, z
#> dbl (1): y
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
#> # A tibble: 3 × 3
#> x y z
#> <chr> <dbl> <chr>
#> 1 <NA> 3 yes
#> 2 NA NA no
#> 3 12 12 NAYou can see from the output tibble that all the missing values are now denoted as NA.
::: {.infobox .caution data-latex=““}
Note that the quotation marks around 999 and -999 are necessary since the na argument is expecting a character vector.
read_csv("x,y,z
999,3,5
999,-999,6", na = c(999, -999))
#> Error in enc2utf8(na): argument is not a character vectorYou can see an error message, stating that it is expecting character values, but got double values.
4.3.4 Importing Data From a Delimited File
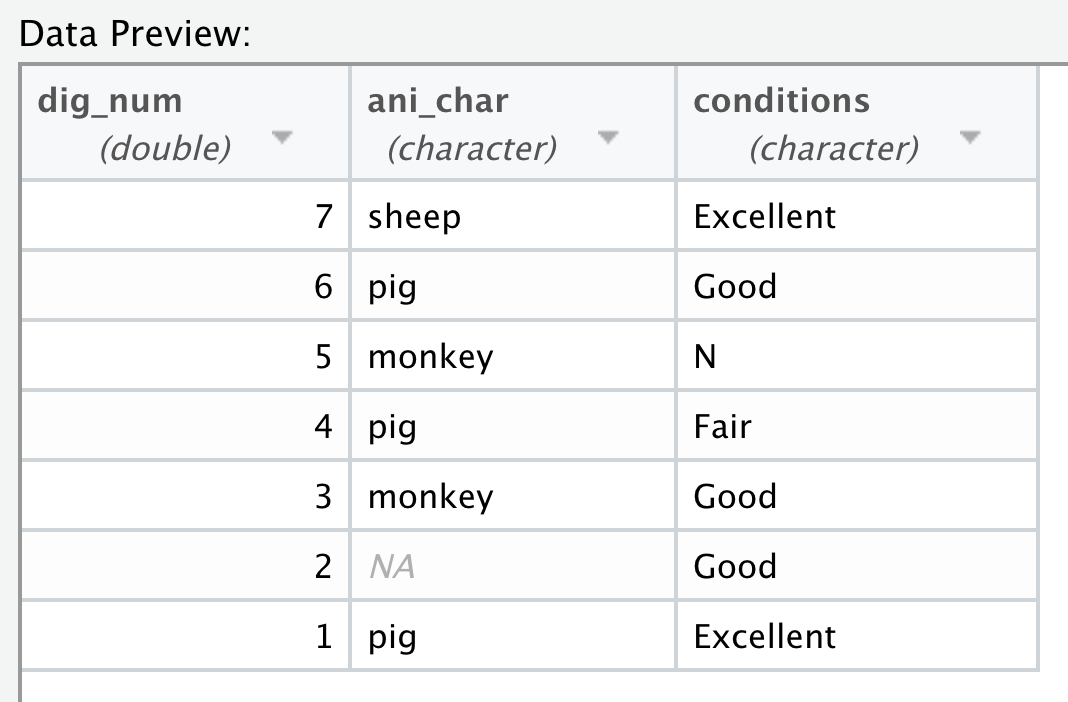
You now know how to import data from a .csv file using the read_csv() function. More generally, read_delim() allows us to import data from a delimited file with any chosen delimiter. The usage of read_delim() is almost identical to read_csv(), except that it has an additional argument delim, which specifies the delimiter to be used. Let’s see the following example with * as the delimiter. Note that we are using the “my_animals_star.csv” file generated in Section 4.2.4.
Let’s try to use read_csv() instead to check what will you get.
read_csv("data/gm_small_star.csv")
#> Rows: 5 Columns: 1
#> ── Column specification ─────────────────────────────────────────
#> Delimiter: ","
#> chr (1): country*year*gender*continent
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
#> # A tibble: 5 × 1
#> `country*year*gender*continent`
#> <chr>
#> 1 Albania*2004*female*Europe
#> 2 Andorra*2004*female*NA
#> 3 United Arab Emirates*2004*female*Asia
#> 4 Argentina*2004*female*Americas
#> 5 Armenia*2004*female*AsiaYou will see that the imported data has only one variable named country*year*HDI since the function will interpret the whole thing as a single variable.
4.3.6 Exercises
For the “my_data_na.csv” file you created in Exercise 2 in Section 4.2, write R code to read the file into an object with name
my_data.First, look at the code below.
Which of the following are the column names of the d1?
X1,X2, andX3x,y, andz
- First, look at the code below.
What will be the column name(s) of the d1?